Pion-LT analysis instructions/adjusting cuts
From HallCWiki
Jump to navigationJump to search
The analysis of the physics data taken in the PionLT experiment is a multi step process -
- First, the raw data is replayed using hcana
- The replay file is then trimmed and cuts are applied using a python script
- /home/cdaq/hallc-online/hallc_replay_lt/UTIL_PION/scripts/online_pion_physics/pion_prod_analysis_sw.py
- A second python script then creates some plots from the trimmed data file
Rough selection cuts are applied to the data at the replay stage. Shift workers should not adjust hcana level cuts unless absolutely neccessary. More refined selection cuts are applied using the first python script, information on how to adjust these cuts is provided below.
Adjusting Analysis Cuts in the PionLT Experiment
- The pion_prod_analysis_sw.py script analyses a replay file and applies selection cuts
- The pion_prod_analysis_sw.py script gets cut definitions from various files in /home/cdaq/hallc-online/hallc_replay_lt/UTIL_PION/DB/CUTS
- The defined cuts automatically get relevant parameters from .csv files located in /home/cdaq/hallc-online/hallc_replay_lt/UTIL_PION/DB/PARAM/
- Parameter values are selected based upon run number (which is provided as an argument to the script when running it)
- Typically, a shift worker is only likely to need (or want) to change timing parameters
- Cut definitions should only be changed in consultation with a PionLT expert
Adjusting Cut Parameters
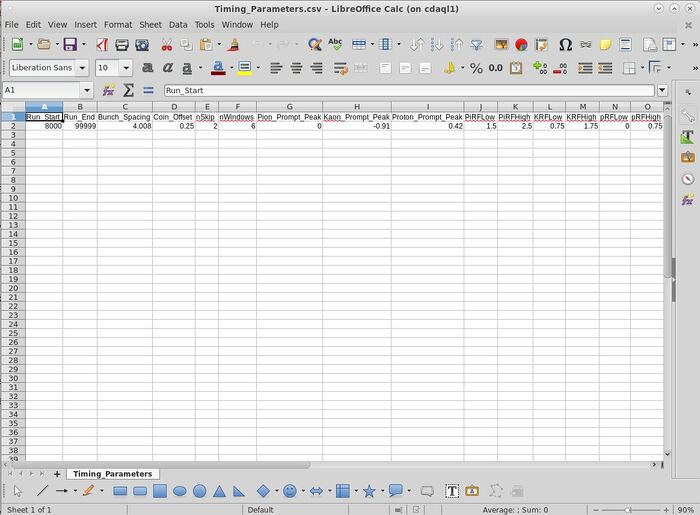
- When adjusting any of the parameter files, I strongly advise that you open the .csv files using libreoffice
- libreoffice /home/cdaq/hallc-online/hallc_replay_lt/UTIL_PION/DB/PARAM/Timing_Parameters.csv
- If you edit anything, make sure you save the file as a .csv file!
- It is unlikely that you will ever need to change any non-timing related parameters
- Consult a PionLT expert first if you need to do so
Timing Cuts
- The "Timing_Parameters.csv" file looks like;
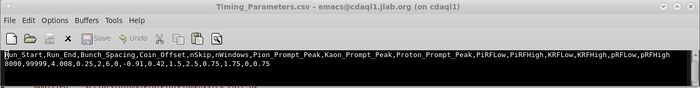
- OR if you didn't take my advice and opened it in emacs, it'll look like this
Coincidence Time Cuts
- Incorrectly set timing parameters may cause the wrong coincidence time peak to be selected
- The most likely culprit for any issues is the "Pion_Prompt_Peak" value
- This should correspond to the prompt peak in the pion coincidence time spectrum
- Bunch_Spacing, Coin_Offset, nWindows and nSKip are all also used to select the prompt and random windows, but these should not be adjusted
- See pages 7 and 8 of pdf produced at the end of the run
- If the value in the .csv file for the run you're analysing does not match what you're seeing on the pdf, edit the value in the .csv file
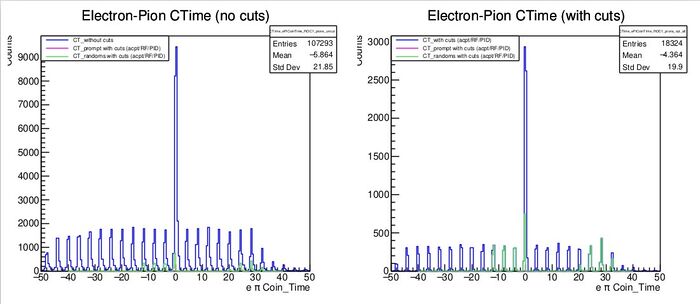
- Shown below is an example of an incorrectly set peak position, and a correctly set one as seen in the plots at the top of page 7
- Here the "Pion_Prompt_Peak" value was set to 10 in the parameter file
- Because it was set poorly, any physics quantities are garbage
- We see only green peaks (corresponding to the random windows), and no magenta peak (the prompt window)
- From the plot, we can clearly see a large peak at 0 which is the correct prompt peak
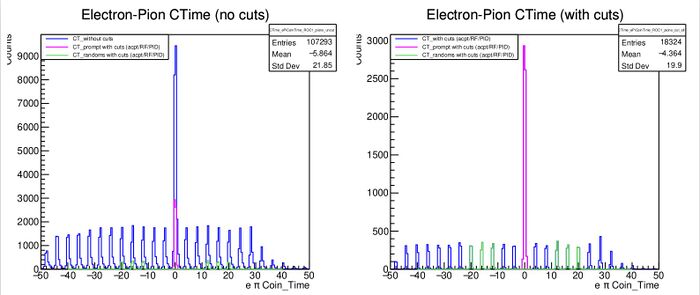
- I went back, set "Pion_Prompt_Peak" to 0 in the csv file for this run and voila, we get the plot below when we reprocess the replay with our two python scripts
- Note that now, we have a magenta peak that matches up perfectly with the prompt peak in the coin time spectrum
- Our physics quantities now look great (take my word for it)
- Note that the prompt peak may not be exactly at 0, you should establish the correct pion prompt peak position from the Electron-Pion CTime (with cuts) plot
RF Timing Cuts
Coming soon - some instructions on setting the RF timing cut window
Notes
- The offset is now a purely hcana level thing (set in standard.kinematics)
- Use the plots on page 4 to determine the "correct" cuts, may not be centred at 0!
- Add image of well set cuts and poorly set cuts
- If unsure, set them wide open (0 to bunch spacing)